About
Extensive research has shown the similarities in human neonates compared to swine neonates regarding neonatal nutrition and digestion as well as the immunoregulatory system thus rendering this type of model preferable over mice or other rodent species for pre-clinical trials. Changes in neonatal pigs represent rapid intestinal growth and functions surrounding nutrient absorption and immune functions. When studying the effects of diseases such as Enteroaggregative E. coli or rotovirus which are particular to neonates, the demand to use a model that is more representative of the chronic infections is necessary. Swine have a gestation length of 113-115 days and synchronization techniques allow for timed parturition and project set-up. Piglets used for neonatal projects are removed from the mother immediately at birth to be placed into highly regulated state of the art facilities uniquely designed for the piglets. Our facilities are able to provide specialty care for early weaned neonates and maintain projects for long term. Piglets have twenty-four hour access to milk by means of our custom designed bottles which are continuously checked by trained personnel. Piglets are housed in their own incubator for intensive monitoring and thermoregulation control as well as individual feeding. Specialty care throughout the duration of their lives is under strict control of IACUC approved protocols.
Selected Publications
- Differential requirements for proliferation of CD4+ and gammadelta+ T cells to spirochetal antigens.
- Hontecillas R, Bassaganya-Riera J. Cell Immunol. 2003 Jul;224(1):38-46.
- Conjugated linoleic acid ameliorates viral infectivity in a pig model of virally induced immunosuppression.
- Bassaganya-Riera J, Pogranichniy RM, Jobgen SC, Halbur PG, Yoon KJ, O’Shea M, Mohede I, Hontecillas R. J Nutr. 2003 Oct;133(10):3204-]14.
- Dietary conjugated linoleic acid modulates phenotype and effector functions of porcine CD8(+) lymphocytes.
- Bassaganya-Riera J, Hontecillas R, Zimmerman DR, Wannemuehler MJ. J Nutr. 2001 Sep;131(9):2370-7.
- Effects of dietary conjugated linoleic acid in nursery pigs of dirty and clean environments on growth, empty body composition, and immune competence.
- Bassaganya-Riera J, Hontecillas-Magarzo R, Bregendahl K, Wannemuehler MJ, Zimmerman DR. J Anim Sci. 2001 Mar;79(3):714-21.
- Arachidonic acid and docosahexaenoic acid-enriched formulas modulate antigen-specific T cell responses to influenza virus in neonatal piglets.
- Bassaganya-Riera J, Guri AJ, Noble AM, Reynolds KA, King J, Wood CM, Ashby M, Rai D, Hontecillas R. Am J Clin Nutr. 2007 Mar;85(3):824-36.
About
Inflammatory bowel disease (IBD) is a chronic reoccurring inflammatory illness affecting millions of people worldwide. IBD has two clinical manifestations: Crohn’s disease and ulcerative colitis. The NIMML developed an acute model of colitis in pigs by intragastric administration of dextran sulfate sodium (DSS) which causes epithelial erosion and subsequent leucocytic infiltration. Another chemical irritant which induces a slightly more severe IBD is 2,4,6-trinitrobenzene sulphonic acid (TNBS) which induces T helper 1 (Th1) and Th17 responses in the colonic mucosa. The NIMML can implement pig projects of DSS, TNBS or bacterial-induced colitis to measure the anti-inflammatory efficacy of new IBD treatments. Gut Inflammation Studies Provide Data for:
- Biomarker discovery
- Therapeutic target discovery
- Bioinformatics analyses
- Immunological assays
- Histopathology services
Selected Publications
- CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Bassaganya-Riera J, Hontecillas R. Clin Nutr. 2006 Jun;25(3):454-65. Epub 2006 May 15.
- CD4+ T-cell responses and distribution at the colonic mucosa during Brachyspira hyodysenteriae-induced colitis in pigs Hontecillas R, Bassaganya-Riera J, Wilson J, Hutto DL, Wannemuehler MJ. Immunology. 2005 May;115(1):127-35.
- Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. Hontecillas R, Wannemeulher MJ, Zimmerman DR, Hutto DL, Wilson JH, Ahn DU, Bassaganya-Riera J. J Nutr. 2002 Jul;132(7):2019-27.
Highlights
- Pigs have greater anatomic, physiologic and immunological similarities to humans than mice, the main animal model used in biomedical research, and therefore have an enhanced translational value.
- The newly developed pig model of H. pylori infection closely mimics what has been reported in human clinical settings.
- Similar to humans, following infection of pigs with H. pylori there was an increase in CD4+ and CD8+ T cells, whereas CD8+ T cells remained unchanged in mouse and gerbil models, the main models used in biomedical research.
- The newly developed pig model allows NIMML to comprehensively and systematically investigate immune responses to H. pylori.
Abstract
Helicobacter pylori infection is the leading cause for peptic ulcer disease and gastric adenocarcinoma. Mucosal T cell responses play an important role in mediating H. pylori-related gastric immunopathology. While induced regulatory T (iTreg) cells are required for chronic colonization without disease, T helper (Th)1 effector responses are associated with lower bacterial loads at the expense of gastric pathology. Pigs were inoculated with either H. pylori strain SS1 or J99. Phenotypic and functional changes in peripheral blood mononuclear cell (PBMC) populations were monitored weekly, and mucosal immune responses and bacterial loads were assessed up to two months post-infection.
Both H. pylori strains elicited a Th1 response characterized by increased percentages of CD4+Tbet+ cells and elevated IFN-γ mRNA in PBMC. A subset of CD8+ T cells expressing Tbet and CD16 increased following infection. Moreover, a significant increase in perforin and granzyme mRNA expression was observed in PBMC of infected pigs indicating a predominant cytotoxic immune response. Infiltration of B cells, myeloid cells, T cells expressing Treg- and Th17-associated transcription factors, and cytotoxic T cells was found in the gastric lamina propria of both infected groups. Interestingly, based on bacterial reisolation data, strain SS1 showed greater capacity to colonize and/or persist in the gastric mucosa compared to strain J99.
This novel pig model of infection closely mimics human gastric pathology and presents a suitable avenue for studying effector and regulatory responses towards H. pylori described in humans.
Summary
To assess whether H. pylori infection affects the expansion of specific T cell subsets, we evaluated the expression of the main transcription factors involved in the regulation of CD4+ T cell phenotype: FOXP3 (iTreg and nTreg), Tbet (Th1) and RORγt (Th17). An elevated proportion of CD4+ T cells expressed the Th1-associated transcription factor Tbet in response to H. pylori (Fig. 1A). We observed a transient single peak at day 7 post-infection followed by a sustained increase in CD4+Tbet+ cells from day 28 to day 49 post-challenge in SS1-infected pigs. The same pattern was found in J99-infected pigs except that the response declined by day 49 post-challenge. The percentage of RORgt+CD4+ T cells was significantly different on days 21 post-infection for SS1 and days 28 and 49 post-infection in J99-infected pigs (Fig. 1C). There was a more notable decline in the expression of the Treg-associated transcription factor FOXP3 in CD4+ T cells from pigs infected with the strain SS1 on day 28 post-infection (Fig. 1D) which mirrored the increase in Tbet and coincided with increased transcripts of IFN-g mRNA in PBMC for both SS1 and J99-infected pigs (Fig. 1B). These results provided the first indication of a predominant Th1 response induced in our experimental model by H. pylori.
CD4+ T cells expressing the T cell lineage specific transcription factors (A) Tbet (Th1), (C) RORγt (TH17) and (D) FOXP3 (iTreg, nTreg) were detected by flow cytometry. IFN-γ mRNA levels in PBMC were measured by qRT-PCR (B). Symbols indicate statistical differences between either the H. pylori J99 (#) or SS1 (*) infected group to the control group and between both infected groups (&), n=8-9, mean±SEM, p≤0.05.
Furthermore, we demonstrate that H. pylori infection results in a significant increase of Tbet expressing cytotoxic CD8b+ T cells (Fig. 2A-B). The shift in Tbet expression was first detected on day 35 post-infection when on average 62% of cells had detectable amounts of this transcription factor. The percentage of CD8b+Tbet+ cells declined thereafter although towards the end of the study, on day 49 post-infection, there were still significant differences between infected and non-infected pigs. A closer analysis revealed the expansion of circulating CD8β+ T cells (Fig. 2C) and an increase of CD16 expression on those circulatingCTL (Fig. 2D) upon infection. Furthermore, CD3-CD8a+ NK cells were significantly increased in blood of SS1-infected pigs on day 35 post-infection (Fig. 2E).
Expression of Tbet was assessed in peripheral CTL (CD8β+) on day 42 post-infection (A) and over time (B). Representative flow cytometry dot blots for non infected and H. pylori J99 and SS1 infected pigs are presented. Numbers indicate percentage of positive cells within the single cell population (A). The number of (C) CTL (CD8β+) and (E) NK cells (CD3-CD8α+), were enumerated in PBMC over time. Numbers of cells per ml of blood were calculated by applying the percentage of immune cells obtained by flow cytometry to the concentration of cells in whole blood. The percentage of CD16 expressing CTL was assessed throughout the study (D). Gene expression levels of perforin (F), granzyme A (G) and granzyme B (H) in PBMC were analyzed over time. Symbols indicate statistical differences between either the H. pylori J99 (#) or SS1 (*) infected group to the control group and between both infected groups (&), n=8-9, mean±SEM, p≤0.05.
In concordance with the observed expansion of circulating cytotoxic T cells, we detected a significant upregulation in the expression of genes involved in the cytotoxic activity of CTL and NK cells, perforin, granzyme A and B (Fig. 2F-H). Overall our data suggest the initial induction of an IFN-g-producing Th1 response orchestrated by the transcription factor Tbet and executed by cytotoxic T cells.
Re-isolation of H. pylori from infected pigs was performed at the end of the study. Overall, H. pylori SS1 was recovered from the stomach of all pigs in that group, while H. pylori J99 could only be re-isolated from 8 out of 12 pigs. When looking at bacterial burden in different regions of the stomach we found that the percentage of re-isolation was consistently higher in the SS1-infected group than in the J99 group with the exception of the fundus-A sub-region which showed similar frequencies for both strains (Fig. 3B). Microscopic changes were present in the stomach of both infected groups and were characterized by significant expansion and development of organized lymphoid aggregates and diffuse leukocytic infiltration. Both strains of H. pylori induced organized lymphoid tissue in the stomach mucosa, which was more predominant in the cardiac region (Fig. 3A).
Representative images were taken from hematoxylin and eosin stained specimens collected from the stomach region cardiac B (CB) and pyloric A (PA) of non infected and infected pigs at 10× magnification (A). H. pylori J99 and SS1 were re-isolated from 6 stomach locations at 2 months post-infection. Re-isolation data is expressed as percentage (B).
In summary, our findings that H. pylori elicits Th1 and CTL responses in a novel pig model correlate well with its role as a facultative intracellular pathogen. Clinical and in vitro studies with human cells provide increasing evidence that cytotoxic immune responses play a crucial role in H. pylori pathogenesis. Our data demonstrates for the first time an increase in circulating CTL and NK cells peaking on day 35 post-infection which coincides with increased IFN-γ gene expression.
The exact role of CD8+ T cells and whether they contribute to the depletion of H. pylori from the gastric mucosa deserves further investigation. Findings from re-isolation and histopathology suggest that CD8+ T cell responses elicited upon infection might be ineffective in the elimination of bacteria but rather contribute to tissue damage. Furthermore, the infiltration of regulatory cells found in the stomach at least partially counteracts proinflammatory responses and contribute to bacterial persistence.
Here, we present the first pig model of H. pylori infection that corroborates in an experimental setting that the predominant Th1 response induced by the bacterium leads to the expansion of cytotoxic cells, including CTL and NK cells. The hallmark of the immune response to H. pylori in humans is the infiltration of Treg cells, neutrophils and Th1 cells. We have been able to reproduce these findings in our pig model, showing infiltration of myeloid cells and FOXP3+ T cells suggesting the presence of Tregs in the gastric mucosa. Furthermore, our model shows a strong systemic Th1 response followed by cytotoxic T cell responses. Similar to H. pylori mediated chronic gastritis in humans, bacteria are able to persist in the pig stomach but at the expense of lesion development. While the role of CD8+ T cells in mouse models of H. pylori has only been studied using immunodeficient mice lacking CD4+ T cells, our pig model provides a more suitable in vivo system to study cytotoxic immune responses towards H. pylori observed in humans and an ideal setting for testing new therapeutic approaches.
Link to the raw data can be found here.
Link to the publication can be found here.
Link to the press release can be found here.
Highlights
- Successfully developed a novel pig model of Helicobacter pylori infection
- Demonstrated for the first time strong cytotoxic CD8+ T cell responses during Helicobacter pylori infection in pigs, further confirming the role of H. pylori as an intracellular pathogen
- Identified the contribution of dysregulated immune responses to gastric inflammatory lesions following Helicobacter pylori infection, particularly by IFNγ-producing Th1 cells and inflammatory macrophages in particular
- Mined large RNAseq datasets from time course studies of macrophage-H. pylori co-cultures
- Identified novel immunological and metabolism genes modulated by H. pylori infection
Background and Epidemiology
Helicobacter pylori are gram-negative, miroaerophilic bacteria responsible for a variety of gastro-duodenal pathologies in the developed and developing world [1]. H. pylori is thought to be indigenous to the human population and is well adapted to colonize and persist in the human stomach. Infection is generally asymptomatic not causing clinical symptoms of those infected people. However, a small percentage of infected individuals will ultimately develop duodenal ulcers and gastric cancer due to the inability of the host immune system to clear the infection.
Pathogenesis
Dozens of bacterial factors are involved in H. pylori molecular pathogenesis (i.e. flagella, urease, catalase, neutrophil-activating protein Nap-A, vacA and cagA). These proteins have revealed many aspects of the relationships between the bacteria, the gastric mucosal surface, and the final outcome of the disease. Two of the most studied virulence factors are vacuolotoxin A (VacA) and the cytotoxin-associated gene A (cagA). Secreted VacA triggers pore formation in the cell membrane, endolysomal trafficking modification, cellular vacuolation, immune cell apoptosis and cell inhibition [2, 3]. CagA is an effector protein injected into the gastric epithelial cells by a type IV secretion system encoded by the cag pathogenicity island (cagPAI). Once inside the host cells, it localizes under the point of bacterial attachment and interacts with the protein zonulin (ZO-1) and the junctional adhesion molecule (JAM) [4]. CagA is then phosphorylated on EPIYA repeats in its phosphotyrosine (PY) region induce secretion of interleukin 8 (IL-8) [5].
Immune response towards H. pylori
As shown in figure 1, the inflammatory response towards H. pylori is initiated through the interaction between the pathogen lipopolysaccharides (LPS) and the Toll-like receptors (TLR) expressed on gastric epithelial cells [6]. Once in the gastric lamina propria, H. pylori is mainly found inside macrophages where their interaction leads to macrophage activation and cytokine release [7]. Macrophages interact with T helper (Th) cells during infection and release cell-polarizing cytokines such as IL-17 [8]. In addition, H. pylori infection also involves neutrophils and increased antigen presenting activity of dendritic cells (DC) [9].
Modeling immunity to H. pylori
MIEP is basically focused on the characterization of the mechanisms underlying immune responses to enteric pathogens by integrating mathematical and computational modeling approaches with experimental data (Figure 2). The integration of modeling and experimental approaches provides unprecedented opportunities for systems-level knowledge discovery. Given the complexity of the host-H. pylori interactions at the systems level and the broad range of possible outcomes, our team has developed a computational model of the gastric mucosal immune response towards H. pylori infection. Currently, the model is able to predict the distinct time-dependent behavior of the three main CD4+ T cells (Th1, Th17 and iTreg) showing an increased Th17 response at the early stage of infection that switches to a Th1 predominance in the chronic phase of the infection.
References
1. Abdulrasheed A, Lawal OO, Abioye-Kuteyi EA, Lamikanra A: Antimicrobial susceptibility of Helicobacter pylori isolates of dyspeptic Nigerian patients. Tropical gastroenterology : official journal of the Digestive Diseases Foundation 2005, 26(2):85-88.
2. Amieva MR, El-Omar EM: Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 2008, 134(1):306-323.
3. Tanaka S, Mizuno M, Maga T, Yoshinaga F, Tomoda J, Nasu J, Okada H, Yokota K, Oguma K, Shiratori Y et al: H. pylori decreases gastric mucin synthesis via inhibition of galactosyltransferase. Hepato-gastroenterology 2003, 50(53):1739-1742.
4.Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S: Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 2003, 300(5624):1430-1434.
5. Eaton KA, Kersulyte D, Mefford M, Danon SJ, Krakowka S, Berg DE: Role of Helicobacter pylori cag region genes in colonization and gastritis in two animal models. Infection and immunity 2001, 69(5):2902-2908.
6. Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS: Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS pathogens 2011, 7(12):e1002454.
7. Ito T, Kobayashi D, Uchida K, Takemura T, Nagaoka S, Kobayashi I, Yokoyama T, Ishige I, Ishige Y, Ishida N et al: Helicobacter pylori invades the gastric mucosa and translocates to the gastric lymph nodes. Laboratory investigation; a journal of technical methods and pathology 2008, 88(6):664-681.
8. Dong C: TH17 cells in development: an updated view of their molecular identity and genetic programming. Nature reviews Immunology 2008, 8(5):337-348.
9. Zhuang Y, Shi Y, Liu XF, Zhang JY, Liu T, Fan X, Luo J, Wu C, Yu S, Chen L et al: Helicobacter pylori-infected macrophages induce Th17 cell differentiation. Immunobiology 2011, 216(1-2):200-207.
Highlights
- NIMML built a mathematical and computational model describing the immune response towards Helicobacter pylori infection.
- NIMML used two different strategies to model these reactions: ODE-based modeling and ABM-based modeling.
- Both strategies resulted in similar results, proving that the combination of ODE and ABM, when used as a complementary approach, can result in the identification of crucial nodes and in the generation of novel hypotheses.
Modeling of immune responses towards H. pylori - Ordinary differential equation model
Given the complexity of the host-H. pylori interaction and to facilitate a better understanding of the gastric mucosal immune response during H. pylori infection, we constructed a computational and mathematical model (Figure 1). The structural network of the model is comprised of three different compartments representing the effector sites: the gastric lumen, the epithelium and the gastric lamina propria plus a fourth inductor compartment representing the gastric lymph nodes (GLN). This network was used for equation-based and agent-based modeling efforts. The equation-based model is comprised by 24 species and 26 ordinary differential equations (ODE) that drive 43 reactions in both gastric mucosa and GLN, and encompasses both inflammatory and regulatory pathways.
Helicobacter Pylori Model
Our computational simulations using ODE modeling show a distinct time-dependent behavior in the three CD4+ T cell phenotypes (i.e., Th1, Th17 and iTreg) represented in the model during H. pylori infection. Whereas Th17 is crucial at an early stage of the infection, Th1 predominates over Th17 and is key for the chronicity of the infection in the gastric LP. Together with these responses, there is a regulatory T cell upregulation peaking at day 30 and being persistent over the infection. Experimentaly, stomachs of H. pylori infected mice have more histopathological lesions(Figure 3F, 3G) To determine and track the main responsible subsets triggering such lesions, sensitivity analysis (SA) methods were applied. Results showed how at the early stage of infection, the epithelial cell damage is mainly caused by the bacterium itself (Figure 2A). Interestingly, as the infection progresses, a trend towards Th1 cells triggering epithelial cell damage (Figure 2B) is observed. At the chronic phase of the infection, results showed a dramatic increase of Th1- and Th17-inducing epithelial cell damage (Figure 2C). Of note, SA performed in the deterministic model at day 60 post infection also showed how Th1 and Th17 in both LP and GLN were contributing to the formation and accumulation of damaged epithelial cells as well as M1 macrophage differentiation, whereas H. pylori exhibited no impact on such formation (Figure 8D).
The following is a list of archived COPASI Helicobacter Pylori computational model releases. Please click on individual releases for more details.
The primary MIEP team members responsible for maintaining this model are Adria Carbo, Mireia Pedragosa and Kate Wendelsdorf at the Virginia Bioinformatics Institute. Please contact them with any questions or comments. For the latest release, The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
- Fourth, the latest, release on May 27th, 2012
- Third release on April 28th, 2012
- Second release on January 14th, 2011
- First release on September 15th, 2011
Modeling of immune responses towards H. pylori - Agent Based model
Modeling H. pylori using ENISI and Cell Designer Updated on December 29th 2012: The ENISI model has been progressed in the following two areas: Sensitivity Analysis and Cell Movement Modeling. To further characterize the immunological mechanisms underlying mucosal immune responses to H. pylori in a stochastic system, we used ABM based on parameter values derived from our ODE model. When probabilistic approaches are used, the complex immunological processes can be better represented. We adopted the ABM tool ENteric Immune Simulator (ENISI) developed by us and available here. In this case, a stochastic Agent Based Modeling approach has been used to better represent the biological system and add a complementary view on the cascade after infection. ABM adds randomness to the biological systems, which can help to better represent complex cellular responses and to take into account the individual behaviors of cells as well as the role spatiotemporal features. Thus, stochastic models can provide novel insights into the effect of cognate and non-cognate interactions, representing entire systems with a greater granularity and capturing cell-cell interactions. By simulating individual behaviors of agents, ABM better represents cross-linked, complex and nonlinear processes with multiple feedback loops and, provides a more comprehensive and interactive modeling of mucosal immune responses to H. pylori. The ability of ABM to encompass multiple scales of biological processes and incorporate spatiotemporal considerations, coupled with an intuitive modeling paradigm, underscores the added value of this modeling framework in translational systems immunology and immunoinformatics research. Given the complexity, nonlinearity and abundance of feedback loops in mucosal immune responses to H. pylori and to facilitate a better understanding of the mechanisms underlying such immune responses at the systems level, we constructed a SBML network model depicting the major effector and regulatory pathways evoked during H. pylori infection (Figure 1). Three different compartments are represented: gastric lumen, epithelium and lamina propria. Effector subsets are highlighted in red whereas regulatory subsets are highlighted in blue. Some updates on the HP ENISI model contain:
- New states of bacteria have been added which incorporates alive, dead and resting bacteria.
- The lifetime of each cell has been modified. Now each cell has a lifetime with normal distribution with a mean and standard deviation.
- We have changed the model parameters to adequate the model to different scenarios of experimental interest. Those scenarios can be accessed here.
- We have reduced some unnecessary states that were not needed in the system.
- We have created a generator application that creates the initial simulation files. This application has been updated with the additional initial states.
The model is available for download in CellDesigner xml format (right click the link or ctrl+left click for Mac machines to save the model source file). We have tested that the model is compatible with Cell Designer 4.2. The primary MIEP team members responsible for maintaining this model are Maksudul Alam, Adria Carbo and Yongguo Mei at the Virginia Bioinformatics Institute. Please contact them with any questions or comments. (Click on the image for a user-friendly interactive CellPublisher model.)

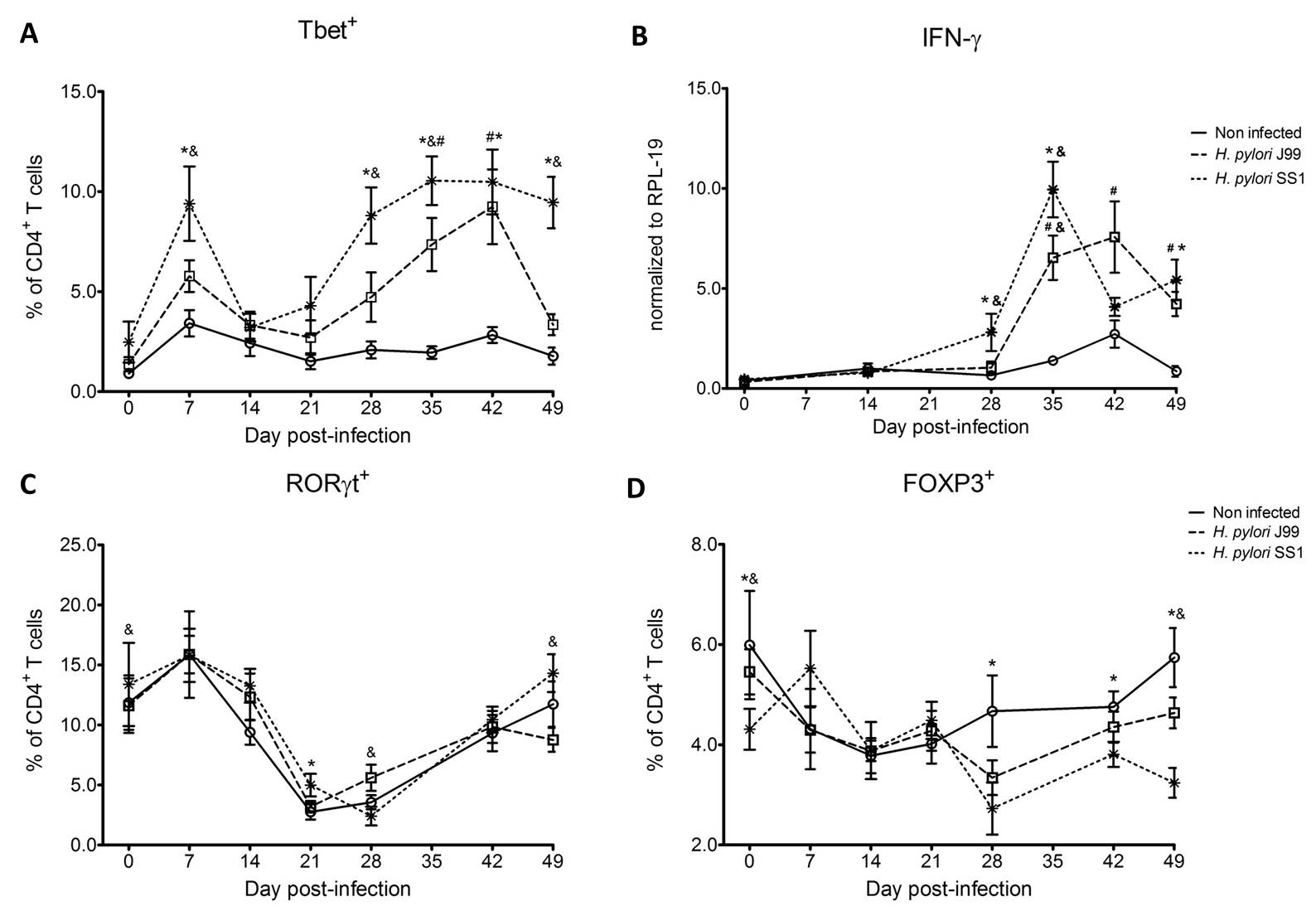 Figure 1. Increase in circulating Th1 cells and IFN-γ expression upon H. pylori infection.
Figure 1. Increase in circulating Th1 cells and IFN-γ expression upon H. pylori infection.
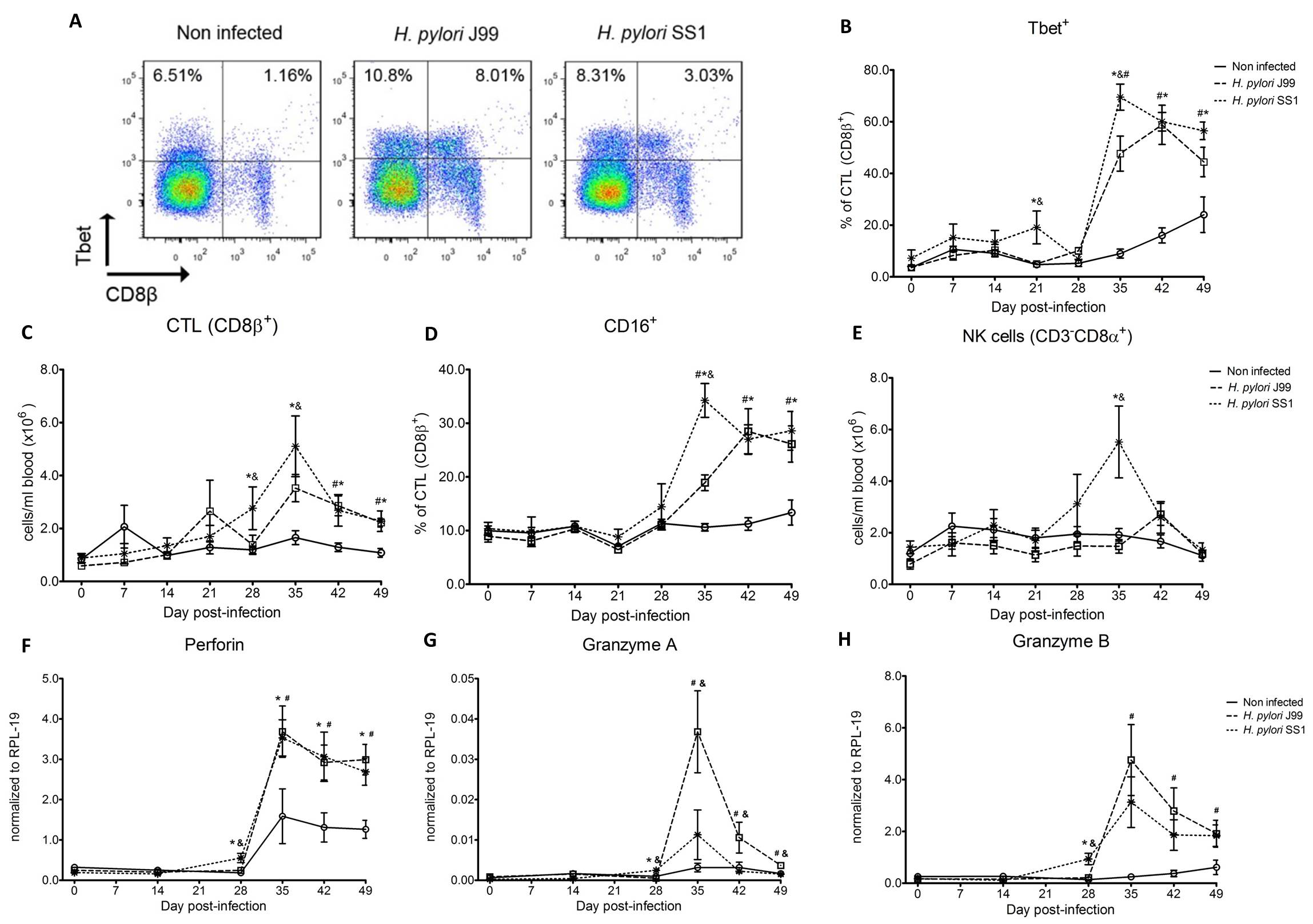 Figure 2. Increased circulating and functional cytotoxic cells upon H. pylori infection.
Figure 2. Increased circulating and functional cytotoxic cells upon H. pylori infection.
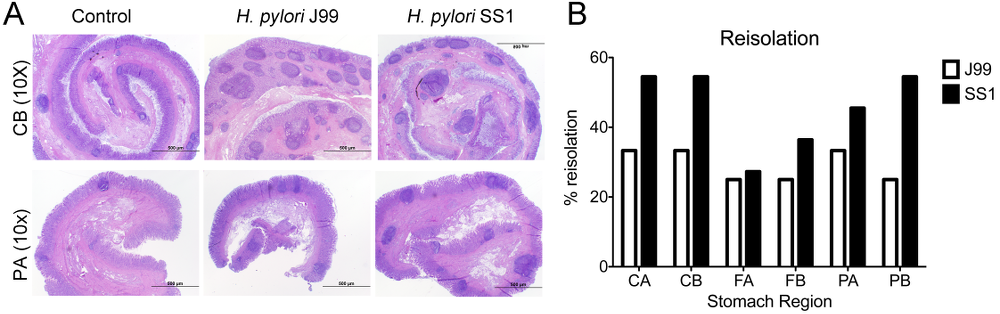 Figure 3. Lesion development upon H.pylori infection and the lack of bacterial clearance upon long-term infection with strain SS1.
Figure 3. Lesion development upon H.pylori infection and the lack of bacterial clearance upon long-term infection with strain SS1.
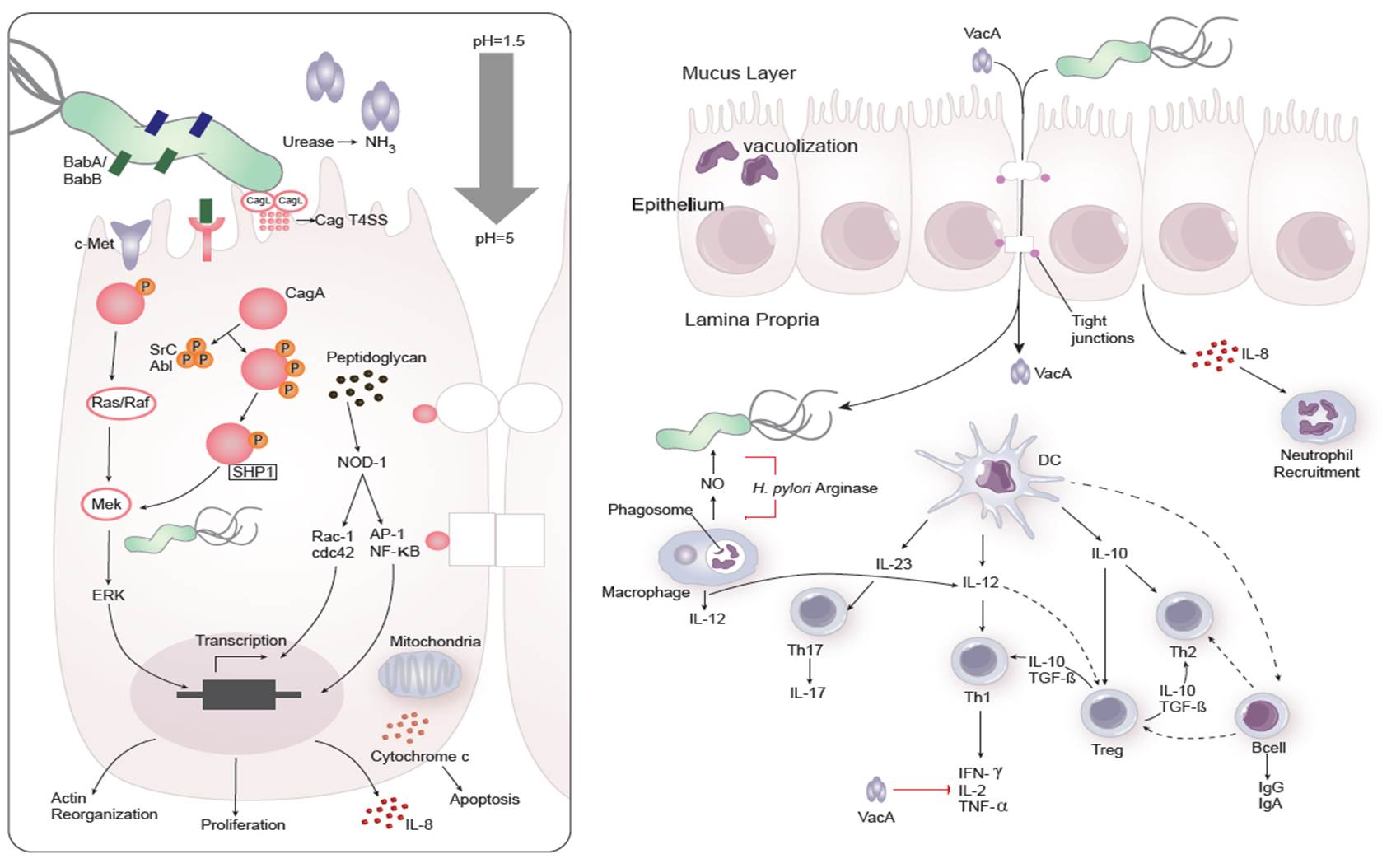 Figure 1. Cellular and molecular pathogenesis of Helicobacter pylori infection
Figure 1. Cellular and molecular pathogenesis of Helicobacter pylori infection
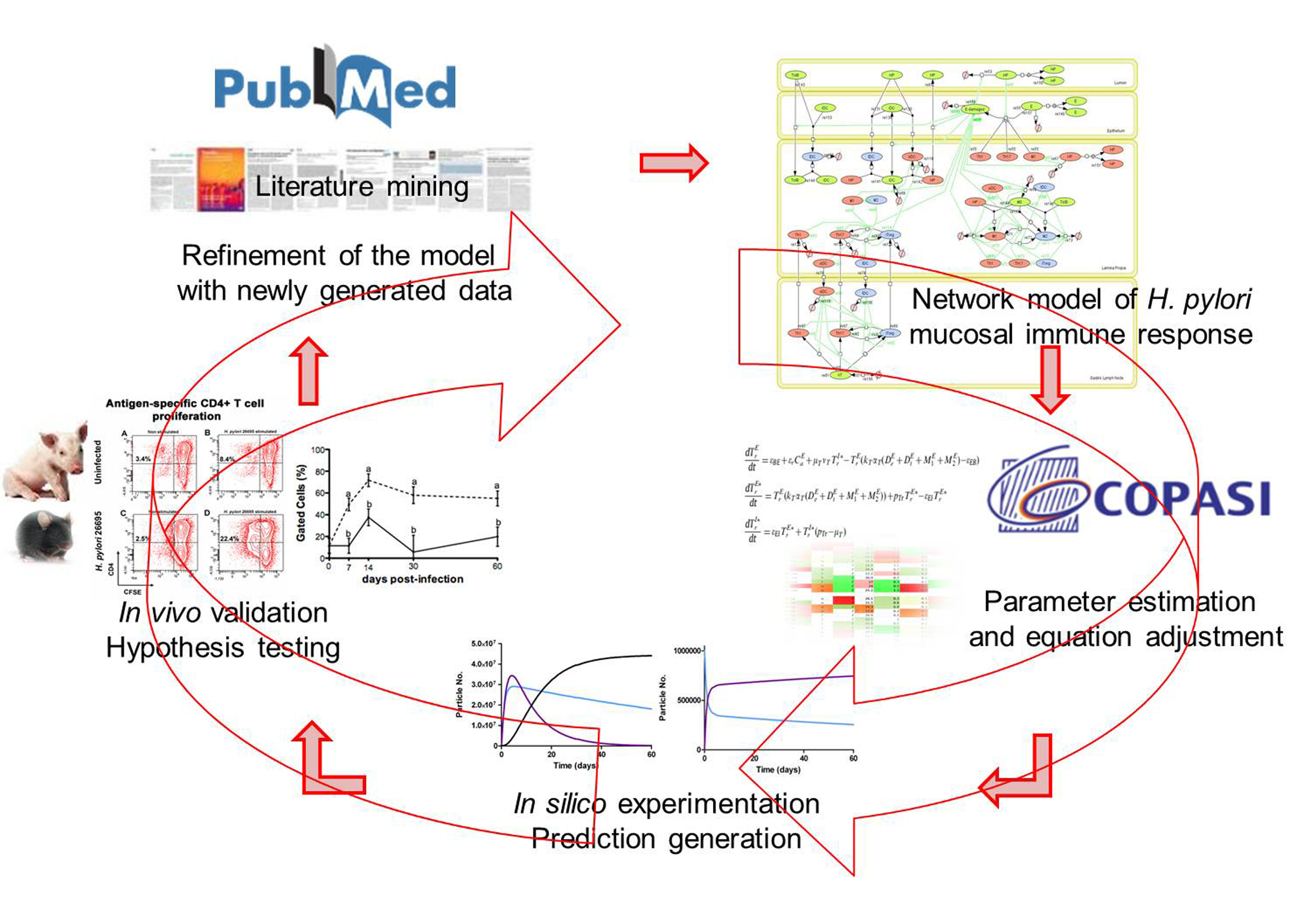 Figure 2. Integration of computational approaches and experimental data.
Figure 2. Integration of computational approaches and experimental data.
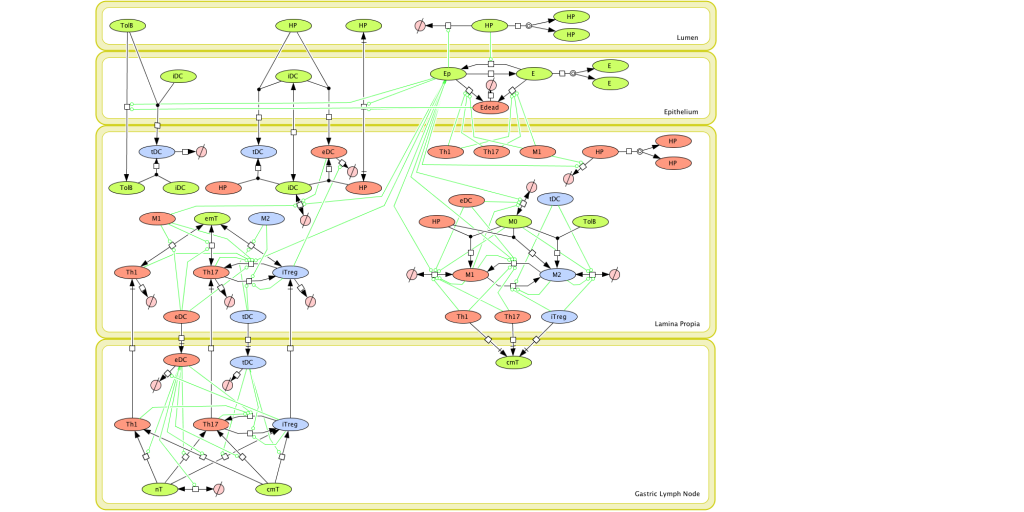 Figure 1. Computational model of the mucosal immune responses to Helicobacter pylori.
Figure 1. Computational model of the mucosal immune responses to Helicobacter pylori.
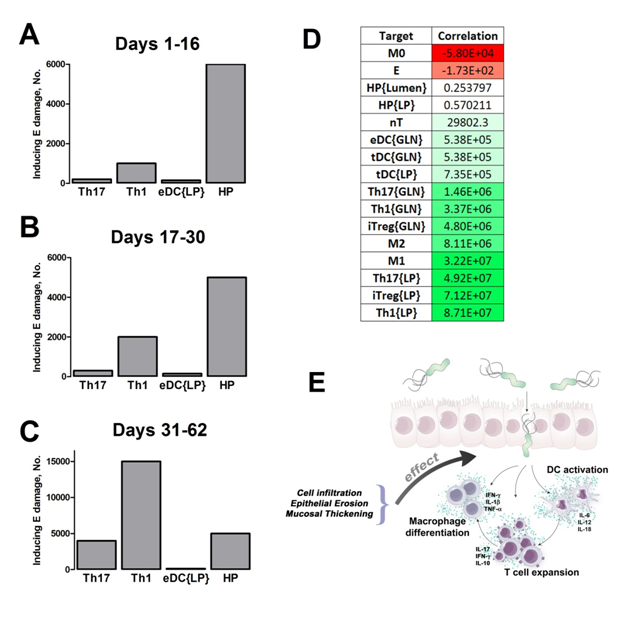 Figure 2. Sensitivity analysis on gastric inflammation lesion formation following Helicobacter pylori infection in silico.
Figure 2. Sensitivity analysis on gastric inflammation lesion formation following Helicobacter pylori infection in silico.
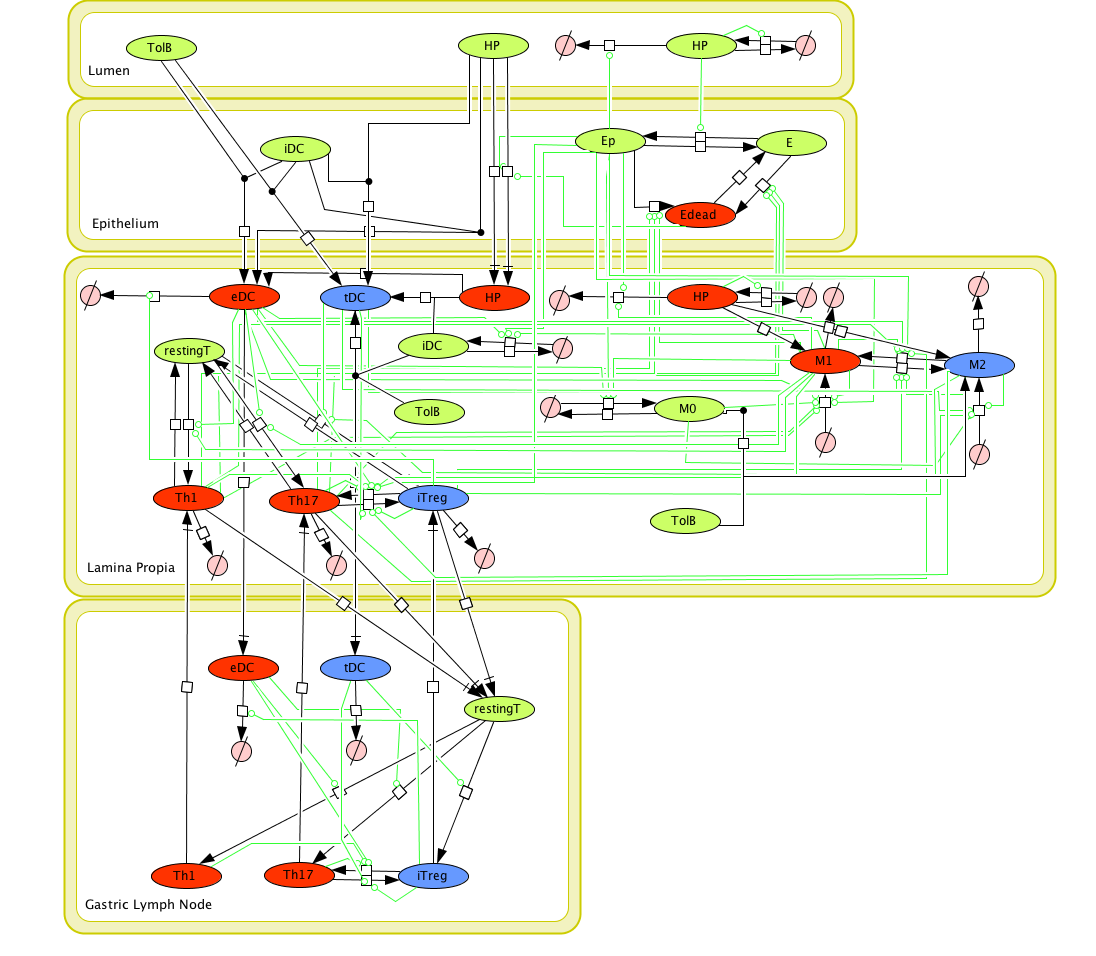 Figure 1. Cell Designer image: ENISI model of H. pylori
Figure 1. Cell Designer image: ENISI model of H. pylori