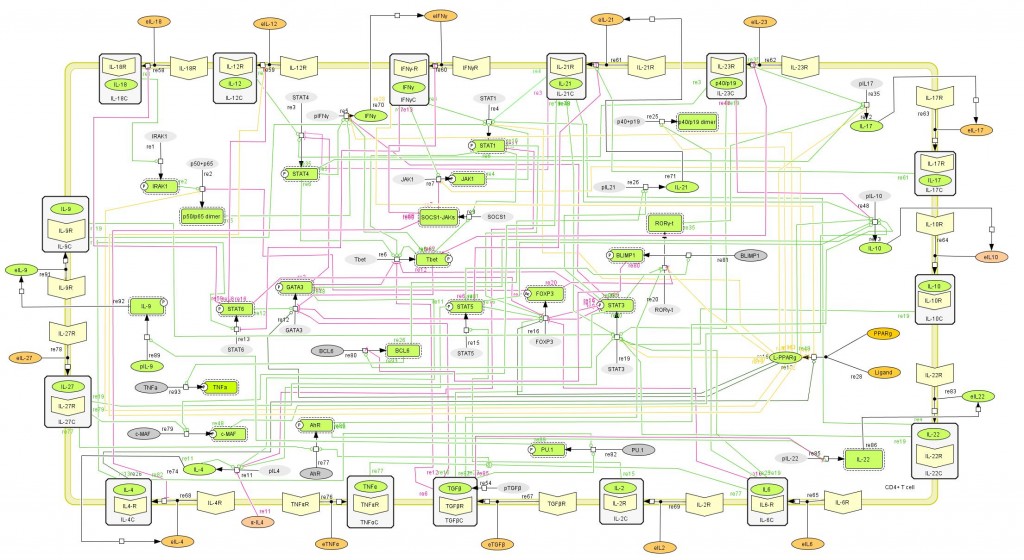
This is a version of MIEP CD4+ T Cell computational model released on Feburary 5th, 2014. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.3.
Here is the release note: The new phenotypes of Th9, Th22, Tr1 and Tfh were added via interactions with new nodes for IL-22, AhR, IL-9, PU.1, BLIMP1, BCL-6, c-MAF, and IL-27. Th9 relies on the presence of IL-4 and TGF-beta in the PU-1 pathway and produces IL-10, IL-9, and IL-21. Cytokines TGF-beta and IL-4 induce the PU.1 transcription factor, which then upregulates IL-9. It also promotes IL-21 via the BCL-6 pathway. IL-21 then promotes c-MAF phosphorylation which upregulates IL-10. Th22 results from the AHR pathway requiring IL-6 and TNF-alpha and upregulates TNF-alpha and IL-22. TNF-alpha and IL-6 work together to promote AhR phosphorylation, which is also promoted by IL-27. AhR then increases IL-6 and IL-22 production. IL-10 promotes the Tr1 phenotype. Tfh results from the BCL-6 pathway and IL-6 and IL-21, and it releases IL-4 and IL-21. STAT-5 promotes BLIMP-1 phosphorylation, which, in turn, downregulates BCL-6. Conversely, BCL-6 is upregulated by STAT-3, which is upregulated by IL-22. IL-22 is promoted by AhR and downregulated by IL-22. BCL-6 then promotes IL-21 and TNF-alpha.
The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
The pathways related to IL-21 have been fully calibrated and in silico experimentation is being run at the moment. Sensitivity analysis on IL-21 has been performed and IL-21 knock-out models have been engineered to better assess the role of IL-21 in CD4+ T cells.
Validation studies have been completed and the manuscript for this study is being put together. Our in vivo and in vitro data validate our in silico prediction, which states that activation of PPAR γ in a fully differentiated Th17 cell triggers a change in transcriptional machinery and function, thus transforming Th17 cells into iTregs.
The IL-21 pathway and the interactions of this cytokine with STAT1 and STAT3 have been re-calibrated with new data. Also, validations studies are being conducted to validate the in silico hypothesis of PPAR γ as a potential candidate in mediating the conversion of Th17 into iTregs after activation. These validation studies consist of in vivo adoptive transfer studies using wild type and PPAR γ CD4-specific null mide as well as in vitro studies. Moreover, parameters for IL-17 internal production have been further refined.
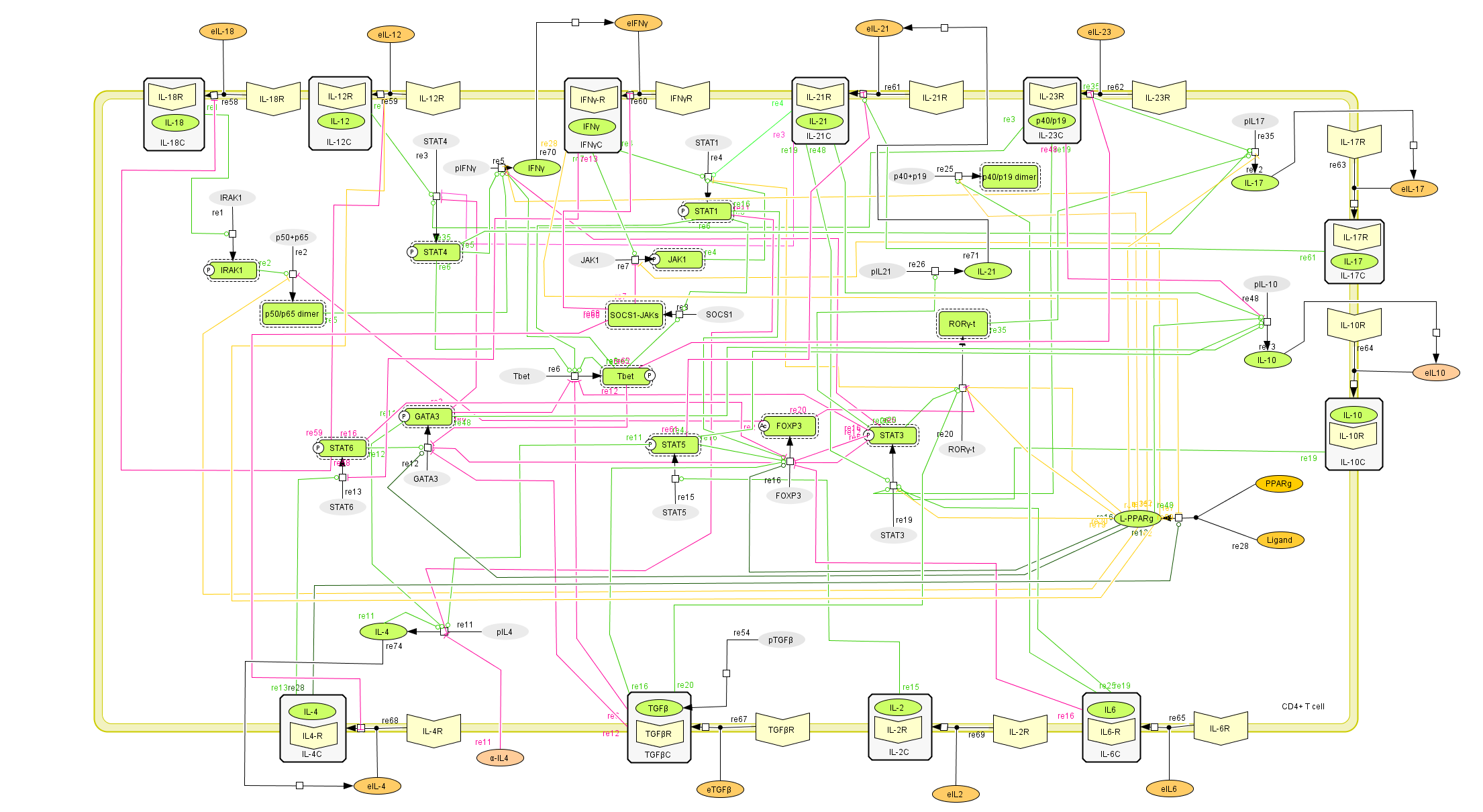
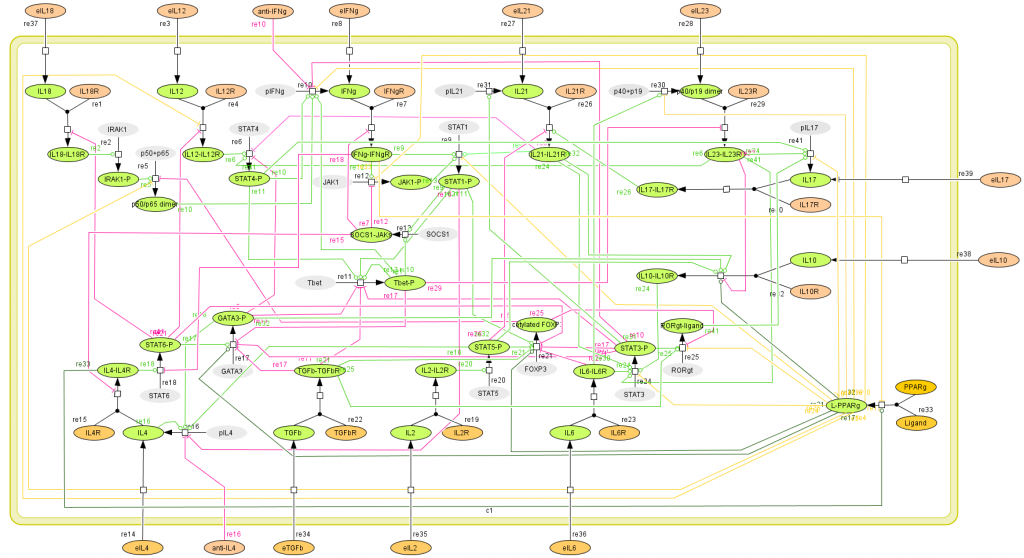
This is a version of MIEP CD4+ T Cell computational model released on January 14th, 2012. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1.
Here is the release note: Cytokine externalization is a key process in CD4+ T cell differentiation because of its paracrine and autocrine effect. The CD4+ differentiation model was before set up with cytokines produced as final products in the mathematical model. With this new update, cytokines are being both internalized and externalized. Thus, a cytokine produced can affect differentiation by being externalized and then act as an inductor. This refinement will allow more complex differentiation studies. Also, activated transcription factors are now highlighted with a discontinued contour to be more easily detectable. Moreover, their shapes have been rearranged so now the transcription factors have a squared shape and the cytokines an oval one.
The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
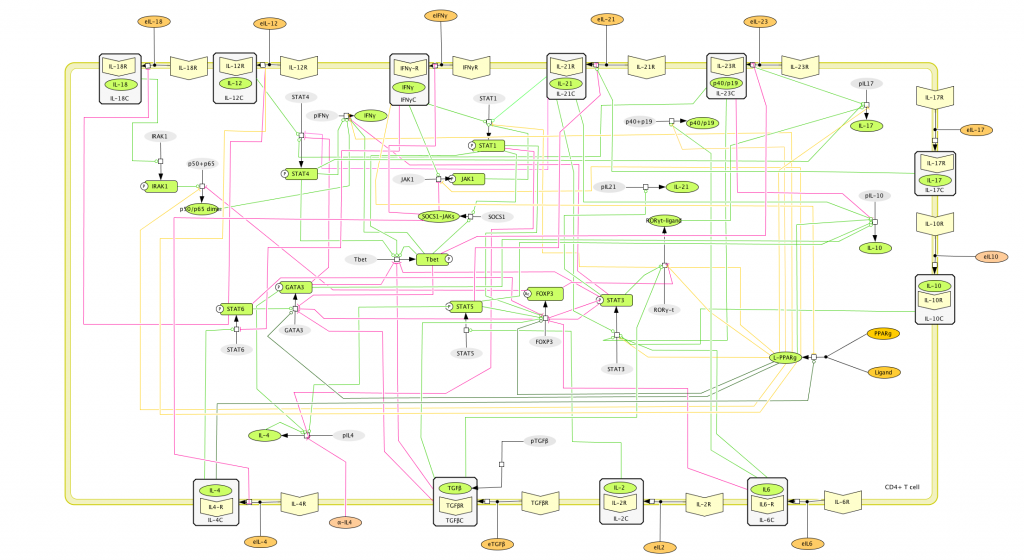
This is an archived version of MIEP CD4+ T Cell computational model released on September 7th, 2011. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
This is an archived version of MIEP CD4+ T Cell computational model released on June 7th, 2011. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
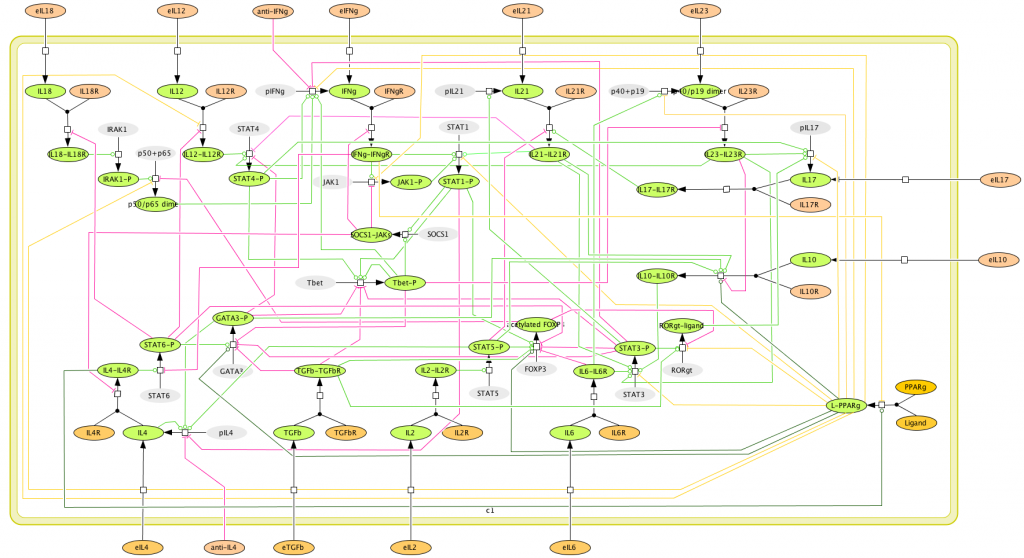
This is an archived version of MIEP CD4+ T Cell computational model released on March 27th, 2011. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.