
ENteric Immune Simulator (ENISI), a multiscale modeling tool for modeling the mucosal immune responses. ENISI can simulate in silico experiments from signaling pathways to tissue level events. ENISI’s architecture integrates multiple modeling technologies. During the development of ENISI, 3 versions were released; ENISE HPC, ENISI Visual, ENISI MSM.

COPASI enables researchers to construct biochemical models that help them understand how a system works. Model parameters can be adjusted as necessary to reproduce experimental results.

CellDesigner is a process diagram editor for gene-regulatory and biochemical networks based on standardized technologies and with wide transportability to other SBML-compliant applications. CellDesigner supports simulation and parameter scan by an integration with SBML ODE Solver, SBML Simulation Core and Copasi.

LabKey is a web-based platform designed for translational research teams. The NIMML uses a customized version of LabKey as its internal electronic notebook and laboratory management information (LIMS) system for real time data-acquisition, data analysis, collaboration, and for publication of raw and processed data

Galaxy captures information that allows users to repeat and understand a complete computational analysis. Galaxy also facilitates sharing and publishing analysis. Galaxy’s public service web site makes analysis tools, genomic data, tutorial demonstrations, persistent workspaces, and publication services available to any scientist.
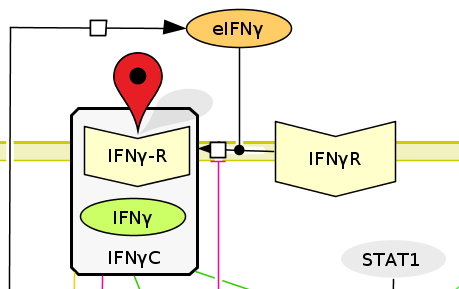
CellPublisher is a free and open source program to make highly interactive representations of biochemical processes. CellPublisher complements CellDesigner by facilitating the understanding of complex diagrams and by providing the possibility to share any CellDesigner diagram online with collaborators and get their feedback. NIMML makes use of CellPublisher to disseminate the models developed to the scientific community in a user friendly way.