COMPUTATIONAL IMMUNOLOGY: Models and Tools. 1st Ed.

This book provides an applied approach to generating and refining hypotheses using computational modeling and simulation studies, and is an ideal resource for immunologists, computational biologists, bioinformaticians, biotechnologists, and computer scientists. The book is based on the notes and presentations stemming from MIEP, a program that integrates computational modeling, big data analytics, portal science and procedural knowledge to engineer synthetic information processing representations of the immune system.
Key Features
- Offers case studies with different levels of complexity
- Provides detailed views useful to experimentalists with limited computational skills
- Explores the usage of simulation for hypothesis generation to accelerate biomedical research
Expected Release Date: 01 Nov 2015
Imprint: Academic Press
Print Book ISBN :9780128036976
Pages:210
Dimensions:235 X 191
For more information, and to order visit Elsevier’s store
Interdisciplinary Program in Genetic Bioinformatics and Computational Biology (GBCB)
Dr. Bassaganya-Riera is an affiliated faculty member of the GBCB program. Bioinformatics and computational biology provide a research platform to acquire, manage, analyze, and display large amounts of data, which in turn catalyze a systems approach to understanding biological organisms, as well as human diseases. Nutritional Immunology students pursuing a GBCB Doctoral degree will integrate the use of bioinformatics tools for solving important problems related to inflammation, immunology, infectious disease or nutritional immunology. The GBCB program provides a training environment that produces a combination of skills in bioinformatics and biosystems that are not commonly found in academia.
Translational Biology, Medicine and Health (TBMH)
The NIMML faculty members are affiliated with the TBMH program. There is an urgent need to address the national need for accelerating the pace of translation of biomedical discoveries into treatment, cures as well as their effective implementation. With translational medicine being an indispensable part of biomedical research, TBMH program is uniquely designed to address this need. NIMML students pursuing a TBMH Doctorate degree will utilize the interdisciplinary approaches; communicate across levels of inquiry by integrating the use of experimental and computational tools in order to solve important problems related to immunology and infectious diseases. The TBMH program provides a training environment for the graduates to take on the future challenges and be prepared for diverse careers that drive today’s biomedical enterprise.
Relevant TBMH Focus Area:
Immunology and Infectious Diseases
http://www.tbmh.vt.edu/focus-areas/immunity-infect/index.html
Graduate Program in Biomedical Engineering (BME)
Dr. Bassaganya-Riera is an affiliated faculty member of the Virginia Tech – Wake Forest School of Biomedical Engineering and Sciences (SBES). SBES is a unique collaboration between the Wake Forest University School of Medicine and Virginia Tech whose purpose is to “provide a framework for the generation and dissemination of knowledge through research and education for the improvement of human and animal health through cooperative advancement in engineering, science and medicine.” Students interested in pursuing the GPI in the Nutritional Immunology Group are encouraged to become BME students so that SBES becomes their academic Department.
The Virginia Bioinformatics Institute (VBI) at Virginia Tech was proud to be hosting the Modeling Mucosal Immunity (MMI) Summer School Program and Symposium on June 9th-13th, 2014. The program was sponsored by VBI’s Center for Modeling Immunity to Enteric Pathogens (MIEP) and was intended for experimental immunologists who wish to gain or expand their understanding of the computational modeling tools used to study immune responses.
“Mathematical and computational models cannot replace experimentation, but they can provide a framework for organizing existing data, generating novel mechanistic hypotheses, and deciding where to focus key validation experiments in time and space. MIEP has built new types of mathematical and computational models that reveal novel mechanisms of immune regulation in the gut mucosa during enteric infection. The MMI Summer School and Symposium will provide a window into such promising computational modeling approaches. Participants will learn how they can use these tools for their own experiments and bring their studies to the next level,” said Dr. Josep Bassaganya-Riera, Professor of Immunology at VBI and Director of MIEP.
In addition to experimental immunologists, computational biology and bioinformatics graduate students at the earliest stages of their studies were also welcome to attend the Summer School. No previous computational modeling experience was required.
WHAT WILL YOU BE LEARNING?
Sunday 8th
Welcome and Installation Gathering
On Sunday, we will host a session in Newman Library to help set up your computer with our university high-speed WiFi connection and the software you will need in the upcoming days. Come and meet your fellow summer school participants!
Monday 9th
COPASI: Computational model creation and simulation
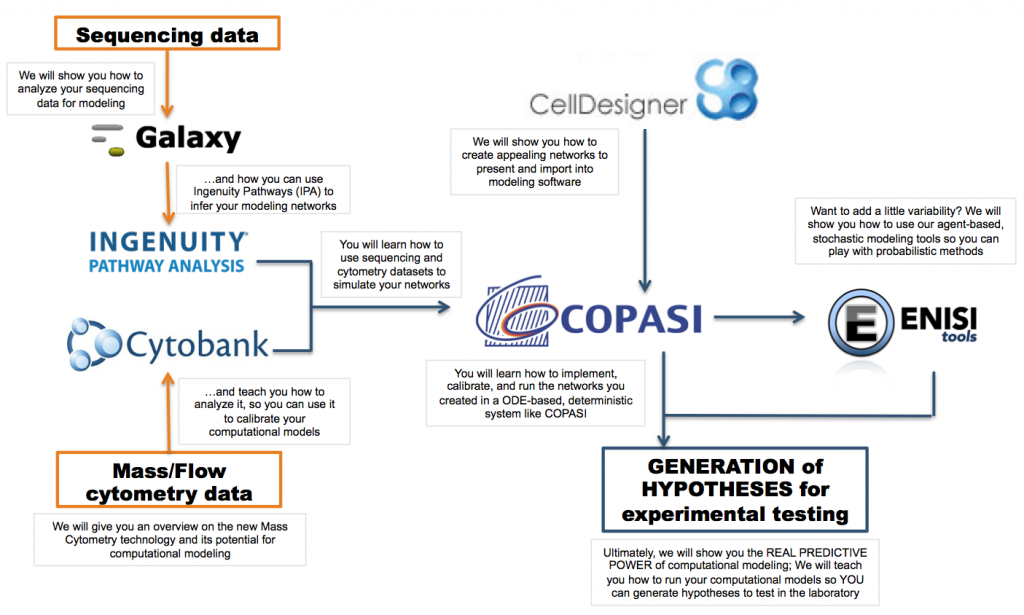
Monday will be centered on the process of model creation and deterministic simulations with ordinary differential equations (ODE). We will create a network from scratch, import it into COPASI, our modeling software, and run simulations to generate hypothesis. We will use our CD4+ T cell differentiation computational model as an example, which will be extensively discussed and presented in the morning.
Tuesday 10th
The ENteric Immune SImulator (ENISI) and Cytobank: continuing with the modeling process
Tuesday will be centered on ENISI, our agent-based modeling tool and how to use different forms of this software. You will also learn how to use Cytobank, a software that analyzes mass cytometry data, and that will help you to generate calibration datasets for modeling purposes.
Wednesday 11th
Computational strategies for Network Inference and Modeling
On Wednesday, we will explore how to analyze RNAseq data and how to use the processed dataset to generate modeling calibration data. We will also show you how to incorporate RNAseq data into IPA, a software used for network inference.
Thursday 12th
Multiscale Modeling: concepts, techniques, and applications
Thursday morning will focus on the concepts, techniques and applications of multiscale modeling. Keynote speakers from the symposium will be present to discuss the latest advances in multiscale modeling. The afternoon will showcase how to integrate all the concepts learnt during the summer school to create computational models and generate hypotheses.
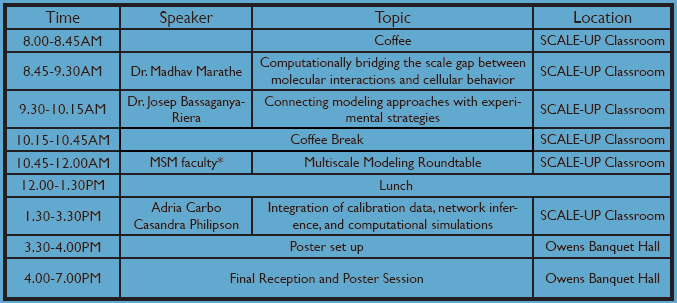
MSM Faculty
To discuss the latets advances in multiscale modeling and debate what the user can do to apply it, we will have a multiscale modeling panel formed by:
- Dr. Josep Bassaganya-Riera (Virginia Tech) *Moderator
- Dr. Martin Meier-Schellerscheim (National Institutes of Health, NIH)
- Dr. Nir Yosef (University of California Berkeley)
- Dr. Madhav Marathe (Virginia Tech)
- Dr. Stuart Sealfon (Mount Sinai Hospital)
- Dr. Doug Lauffenburger (Massachusets Institute of Technology, MIT)
Final Reception & Poster Session
To celebrate the end of the Summer School we will be holding a Poster Session in Owens Banquet Hall. This will be a great opportunity for network and discussion as we will learn a little more about the research being held by other participants of the school. Refreshments and drinks will be provided.

Dr. Allen is an Assistant Professor at the Biomedical Sciences and Pathobiology Department at VMRCVM. His research interests are epithelial cell immunology, host pathogen interactions, and animal models of innate immunity and also allergic disease.

Dr. Bassaganya-Riera directs the Nutritional Immunology and Molecular Medicine Laboratory and is a Professor of Immunology at Virginia Tech. He is the Director of the Modeling Immunity to Enteric Pathogens (MIEP) that investigates the immunoregulatory mechanisms underlying host responses to gut pathogens by applying mathematical systems to mucosal immunology.

Dr. Bevan is the Education Lead in the Modeling Immunity to Enteric Pathogens Center. He is also a Professor at Virginia Tech and his laboratory involves the application of computational molecular modeling to relate the structure and dynamics of molecular systems to function.

Adria is a senior Ph.D. Candidate in the Nutritional Immunology and Molecular Medicine Laboratory. He has been working with the Center for Modeling Immunity to Enteric Pathogens since 2010 and leads the efforts on the CD4+ T cell differentiation modeling.

Dr. Hontecillas is the co-director in the Nutritional Immunology and Molecular Medicine Laboratory and the Immunology Lead and co-director in the Modeling Immunity to Enteric Pathogens Center, where she investigates the immunomodulatory mechanisms of action during Helicobacter pylori, Clostridium difficile, and Escherichia coli infection.

Dr. Hoops is a Bioinformatician at the Nutritional Immunology and Molecular Medicine Laboratory and the developer of COPASI. His research interests are modeling and simulation of biochemical systems as well as management and analysis of systems biology data sets.

Dr. Lauffenburger is a Professor of biological and chemical engineering at the Massachussets Institute of Technology (MIT). He also serves as the Head in the department of Biological Engineering. His research focuses on elucidating important aspects of receptor-mediated regulation of mammalian blood and tissue cell behavioral functions such as proliferation, adhesion, migration, and macromolecular transport.

Pawel Michalak’s research program focuses on comparative genomics, transcriptomics, bioinformatics, and evolution. Recent advances in genomic technologies and bioinformatics, including ‘next-gen’ sequencing of entire genomes and transcriptomes, provide a unique opportunity to investigate how morphological complexity and disease elaborate from genomes through gene regulatory networks. He has been involved in the Bioinformatics aspects of the MIEP project for the last 4 years and has maintained productive research collaborations with the team since then.

Dr. Marathe is the deputy director in the Network Dynamics and Simulation Science Laboratory (NDSSL) at Virginia Tech. He is also the lead in the Modeling Core in the Center for Modeling Immunity to Enteric Pathogens and his research interests are High-Performance Computing (HPC), linked to Agent-Based Modeling (ABM) in the context of infectious diseases.

Dr. Mei is a Senior Software Engineer and Research Scientist at the Modeling Immunity to Enteric Pathogens Center. His interests are developing user-friendly modeling tools for the study of the immune responses to certain pathogens. He is one of the key developers of the ENISI agent-based modeling tool.

Dr. Oestreich is an Assistant Professor at the Virginia Tech Carilion Research Institute and at the Biomedical Sciences and Pathobiology Department at VMRCVM. His laboratory focuses on defining the mechanisms by which transcription factors contribute to cellular diffeentiation and function in the immune system.

Casandra is a Ph.D. student in the Nutritional Immunology and Molecular Medicine Laboratory. Her research involves elucidating the immune mechanisms of action towards infection with Escherichia coli. Furthermore, she has been working on computational models of epithelial cell response.

Dr. Sealfon is a neurologist in the Mount Sinai Hospital in New York and a Professor of Neurology, Pharmacology and Systems Therapeutics, and Neuroscience. He also serves as a chair in the Neurology department. Furthermore, he directs the Program for Research on Immune Modeling and Experimentation (PRIME). His research seeks to develop easy-to-use, predictive mathematical models to better understand patterns of infection among individuals affected by the flu virus.

Dr. Wendelsdorf is a scientist at Ingenuity Pathways (IPA) with the focus on the use of modern software for network inference and analysis. Her modeling background was centered on analyzing large gene expression data sets to identify discrete response states of immune cells.

Dr. Yosef is an Assistant Professor at at the Department of EECS and a member of the Center for Computational Biology at UC Berkeley. The goal of his research is to utilize high-throughput genomic data sets in order to build models that explain how gene expression is regulated.

Monica is a Ph.D. candidate in the Nutritional Immunology and Molecular Medicine Laboratory. Her research is centered on understanding the mechanisms of action towards infection with Clostridium difficile. Her research also involves the analysis of RNA sequencing data with Galaxy.

1. Overview CellDesigner and 3-node network construction
2. Overview of COPASI
3. 3node COPASI
4. Construction 9-node CellDesigner
5. Parameter estimation
6. 9node COPASI voice
7. Overview of the full CD4+ T cell model
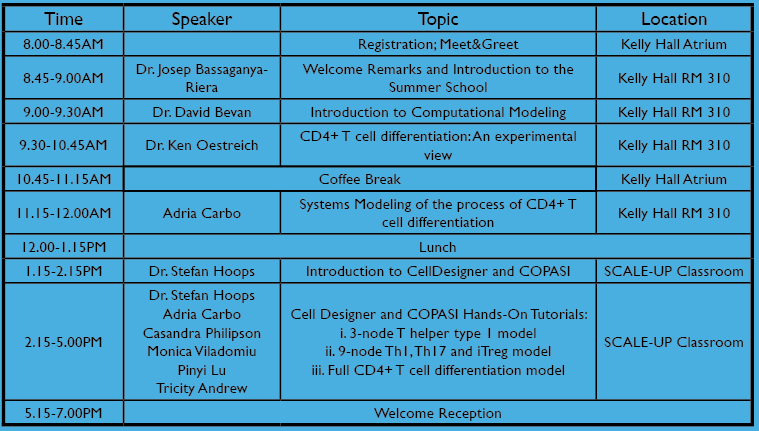
Monday 9th
| Presenter | Topic | Media |
|---|---|---|
| Josep Bassaganya-Riera | Welcome Remarks and Introduction to the Summer School | |
| David Bevan | Introduction to Computational Modeling | |
| Ken Oestreich | CD4+ T cell differentiation: An experimental view | |
| Adria Carbo | PPARγ activation drives Th17 cells into a Tregphenotype: The CD4+ T cell computational model | |
| Stefan Hoops | Introduction to CellDesigner and COPASI | |
| Stefan Hoops, Adria Carbo, Casandra Philipson, Monica Viladomiu, Pinyi Lu, Tricity Andrew | Cell Designer and COPASI Hands-On Tutorials |
|
| Welcome Reception |
|
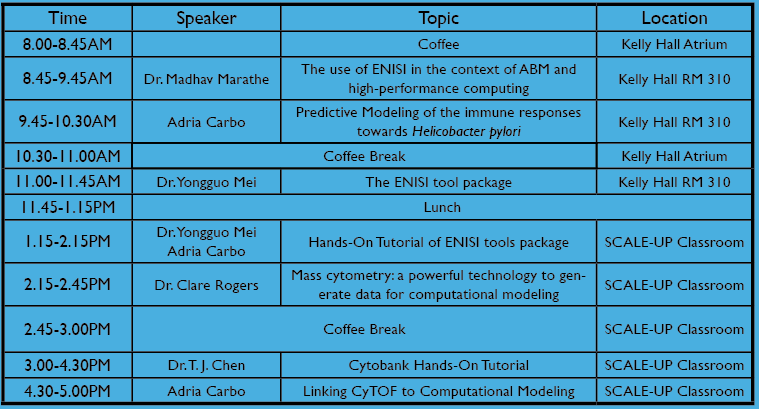
Tuesday 10th
| Presenter | Topic | Media |
|---|---|---|
| Stephen Eubank | The Use of ENISI in the Context of Agent-Based Modeling and High-Performance Computing | |
| Adria Carbo | Predictive Modeling of the immune responses towards Helicobacter pylori | |
| Yongguo Mei | ENISI Tool Suite | |
| Afternoon Session | ||
| Yongguo Mei, Adria Carbo | Hands-On Tutorial of ENISI tools package |
|
| Claire Rogers | Mass cytometry: a powerful technology to generate data for computational modeling |
|
| T.J. Chen | Cytobank Hands-On Tutorial |
|
| Adria Carbo | Linking CyTOF to Computational Modeling |
|
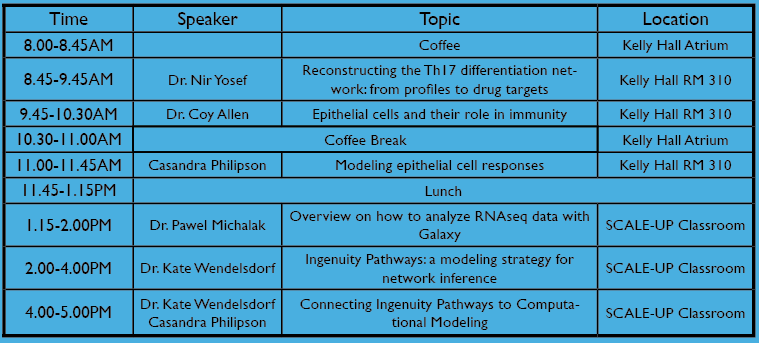
Wednesday 11th
| Presenter | Topic | Media |
|---|---|---|
| Nir Yosef | Reconstructing the Th17 differentiation network: from profiles to drug targets | |
| Coy Allen | Epithelial cells and their role in immunity | |
| Casandra Philipson | Mathematical and Computational Modeling of Epithelial Cell Networks | |
| Pawel Michalak | RNA-seq Analysis in Galaxy | |
| Kate Wendelsdorf | Ingenuity Pathways: a modeling strategy for network inference | |
| Kate Wendelsdorf, Casandra Philipson | Connecting Ingenuity Pathways to Computational Modeling |
Thursday 12th
| Presenter | Topic | Media |
|---|---|---|
| Madhav Marathe | Multi-Scale Modeling in Systems Biology | |
| Josep Bassaganya-Riera | Connecting Data to Models | |
| MSM faculty | Multiscale Modeling Roundtable | |
| Adria Carbo, Casandra Philipson | Integration of calibration data, network inference, and computationial simulations |
|
| Final Reception and Poster Session |
Friday 13th
| Presenter | Topic | Media |
|---|---|---|
| Josep Bassaganya-Riera | Introduction to the Symposium on Computational Immunology | |
| Doug Lauffenburger | In Vivo Systems Analysis of Inflammatory Pathophysiology | |
| Chris Barrett | Computational Modeling and Information Biology | |
| Stuart Sealfon | Information Coding of Dendritic Cell Responses to Virus Infection | |
| Shayne Peirce | Multiscale models for understanding regulators of tissue growth and remodeling | |
| Tom Kepler | Computational Immunology in Vaccine Development | |
| Poster Awards | ||
| Josep Bassaganya-Riera | Center for Modeling Immunity to Enteric Pathogens |

Modeling Mucosal Immunity —— Summer School & Symposium in Computational Immunology
Above is a clip of Tori Godfrey, of the undergraduate students involved in the NIMML team through the Undergraduate Scholars Program (USP) mentioned below. “Summer Academy is a program where new hokies are able to get a head start to their education, whether they be incoming transfer students or freshmen. I am currently a peer mentor for the program and I am participating in undergraduate research here at NIMML. I hope to be a good influence on the students and show them how exciting and stimulating research can be!” - Tori
Research Programs for Undergraduates
- NIMML Undergraduate Scholars Program (USP): In keeping with the Mission, Vision and Values of the Nutritional Immunology and Molecular Medicine Laboratory (NIMML), the NIMML is hosting an Undergraduate Scholars Program that will provide undergraduate students the opportunity to gain valuable research experience and/or course credit training under the direction of experienced transdisciplinary scientists. The program will equip undergraduate students with the research and/or laboratory skills that cannot be gained merely in a classroom setting, while immersing students in cutting edge science and technology environments. Students interested in pursuing graduate degrees will find this type of research experience invaluable in the process of further refining and developing their career interests.

- Scieneering Program: Building on VT’s strengths in science and engineering, the Office of Undergraduate Research, through funding from a prestigious Howard Hughes Medical Institute Science Education Grant, offers a novel and innovative new program focused on interdisciplinary undergraduate studies and research, aptly named Scieneering. Science and engineering sophomores and juniors selected to become Scieneers participate in coursework leading to minors in Interdisciplinary Engineering and Science or Science, Engineering and Law and conduct at least 3 credit- hours of interdisciplinary research mentored by faculty outside their major discipline. This is a great opportunity for students to gain experience tackling real-world, multi-dimensional problems under the direction of a qualified mentor. Scieneers receive a stipend and funds to purchase supplies pertinent to their selected research project.
- Undergraduate Research Program In Nutritional Immunology: The scientific focus of this program for rising juniors in Systems Biology is to integrate transdisciplinary nutritional immunology research with a system’s approach for developing novel therapies against chronic inflammatory diseases. The student is expected to learn acquire a conceptual framework in the interface between nutrition, immunology and systems biology by designing and conducting a research project, becoming familiar with state-of-the-art analytical immunology techniques used to address the working hypotheses, and research data management and analyses. This program is a partnership between the Nutritional Immunology Group at VBI and the Department of Systems’ Biology at the University of Vic (Catalonia, Spain): http://www.uvic.cat.